Digging through some Folding@Home data
Published:
Learning cheminformatics from some Folding@Home data
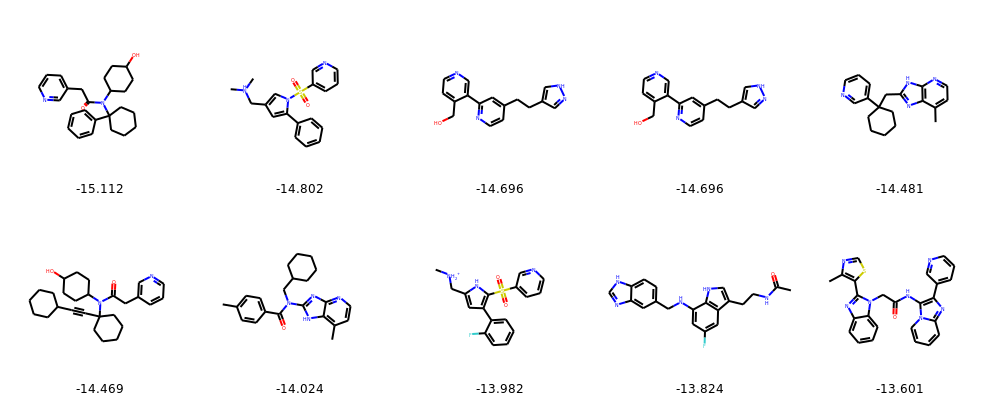
Top 10 (based on Hybrid2 docking score) small molecules
2020-05-06 - 2020-05-11
I have no formal training in cheminformatics, so I am going to be stumbling and learning as I wade through this dataset. I welcome any learning lessons from experts.
This will be an ongoing foray
Source: https://github.com/FoldingAtHome/covid-moonshot
Introduction
Folding@Home is a distributed computing project - allowing molecular simulations to be run in parallel across thousands of different computers with minimal communication. This, combined with other molecular modeling methods, has yielded a lot of open data for others to examine. In particular, I’m interested in the docking screens and compounds targeted by the F@H and postera collaborations
import pandas as pd
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
pd.options.display.max_columns = 999
moonshot_df = pd.read_csv('moonshot-submissions/covid_submissions_all_info.csv')
moonshot_df.head()
| SMILES | CID | creator | fragments | link | real_space | SCR | BB | extended_real_space | in_molport_or_mcule | in_ultimate_mcule | in_emolecules | covalent_frag | covalent_warhead | acrylamide | acrylamide_adduct | chloroacetamide | chloroacetamide_adduct | vinylsulfonamide | vinylsulfonamide_adduct | nitrile | nitrile_adduct | MW | cLogP | HBD | HBA | TPSA | num_criterion_violations | BMS | Dundee | Glaxo | Inpharmatica | LINT | MLSMR | PAINS | SureChEMBL | PostEra | ORDERED | MADE | ASSAYED | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CCN(Cc1cccc(-c2ccncc2)c1)C(=O)Cn1nnc2ccccc21 | AAR-POS-8a4e0f60-1 | Aaron Morris, PostEra | x0072 | https://covid.postera.ai/covid/submissions/AAR... | Z1260533612 | FALSE | FALSE | FALSE | False | False | False | False | False | False | False | False | False | False | False | False | False | 371.444 | 3.5420 | 0 | 5 | 63.91 | 0 | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | True | False | False |
| 1 | O=C(Cn1nnc2ccccc21)NCc1ccc(Oc2cccnc2)c(F)c1 | AAR-POS-8a4e0f60-10 | Aaron Morris, PostEra | x0072 | https://covid.postera.ai/covid/submissions/AAR... | Z826180044 | FALSE | FALSE | s_22____1723102____13206668 | False | False | False | False | False | False | False | False | False | False | False | False | False | 377.379 | 3.0741 | 1 | 6 | 81.93 | 0 | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | True | False | False |
| 2 | CN(Cc1nnc2ccccn12)C(=O)N(Cc1cccs1)c1ccc(Br)cc1 | AAR-POS-8a4e0f60-11 | Aaron Morris, PostEra | x0072 | https://covid.postera.ai/covid/submissions/AAR... | FALSE | FALSE | FALSE | FALSE | False | False | False | False | False | False | False | False | False | False | False | False | False | 456.369 | 4.8119 | 0 | 5 | 53.74 | 0 | PASS | PASS | PASS | Filter9_metal | aryl bromide | PASS | PASS | PASS | PASS | True | False | False |
| 3 | CCN(Cc1cccc(-c2ccncc2)c1)C(=O)Cc1noc2ccccc12 | AAR-POS-8a4e0f60-2 | Aaron Morris, PostEra | x0072 | https://covid.postera.ai/covid/submissions/AAR... | Z1260535907 | FALSE | FALSE | FALSE | False | False | False | False | False | False | False | False | False | False | False | False | False | 371.440 | 4.4810 | 0 | 4 | 59.23 | 0 | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | True | False | False |
| 4 | O=C(NCc1noc2ccccc12)N(Cc1cccs1)c1ccc(F)cc1 | AAR-POS-8a4e0f60-3 | Aaron Morris, PostEra | x0072 | https://covid.postera.ai/covid/submissions/AAR... | FALSE | FALSE | FALSE | s_272164____9388766____17338746 | False | False | False | False | False | False | False | False | False | False | False | False | False | 381.432 | 4.9448 | 1 | 4 | 58.37 | 0 | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | True | False | False |
The moonshot data has a lot of logging/metadata information, some one-hot-encoding information about functional groups, and some additional columns about Glaxo, Dundee, BMS, Lint, PAINS, SureChEMBL - I’m not sure what those additional coluns mean, but the values are binary values, possibly the results of some other test or availability in another databases.
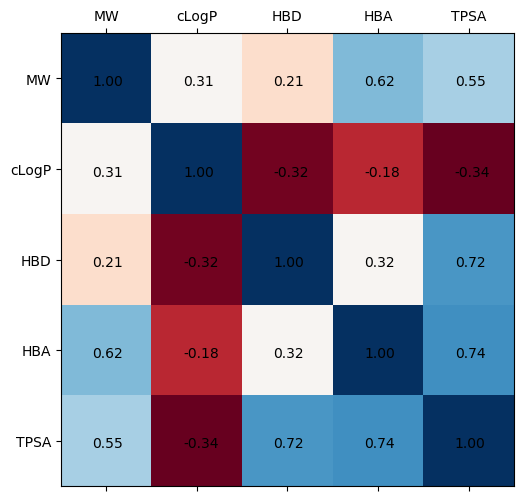
I’m going to focus on the molecular properties: MW, cLogP, HBD, HBA, TPSA
- MW: Molecular Weight
- cLogP: The logarithm of the partition coefficient (ratio of concentrations in octanol vs water, $\log{\frac{c_{octanol}}{c_{water}}}$)
- HBD: Hydrogen bond donors
- HBA: Hydrogen bond acceptors
- TPSA: Topological polar surface area
Some of the correlations make some chemical sense - heavier molecules have more heavy atoms (O, N, F, etc.), but these heavier atoms are also the hydrogen bond acceptors. By that logic, more heavy atoms also coincides with more electronegative atoms, increasing your TPSA. It’s a little convoluted because TPSA looks at the surface, not necessarily the volume of the compound; geometry/shape will influence TPSA. There don’t appear to be any strong correlations with cLogP. Partition coefficients are a complex function of polarity, size/sterics, and shape - a 1:1 correlation with a singular, other variable will be hard to pinpoint
This csv file doesn’t have much other numerical data, but maybe some of those true/false, pass/fail data might be relevant…but I definitely need more context here
fig, ax = plt.subplots(1,1, figsize=(8,6), dpi=100)
cols = ['MW', 'cLogP', 'HBD', 'HBA', 'TPSA']
ax.matshow(moonshot_df[cols].corr(), cmap='RdBu')
ax.set_xticks([i for i,_ in enumerate(cols)])
ax.set_xticklabels(cols)
ax.set_yticks([i for i,_ in enumerate(cols)])
ax.set_yticklabels(cols)
for i, (rowname, row) in enumerate(moonshot_df[cols].corr().iterrows()):
for j, (key, val) in enumerate(row.iteritems()):
ax.annotate(f"{val:0.2f}", xy=(i,j), xytext=(-10, -5), textcoords="offset points")
Some docking results
Okay here’s a couple other CSVs I found, these include some docking scores
- Repurposing scores: “The Drug Repurposing Hub is a curated and annotated collection of FDA-approved drugs, clinical trial drugs, and pre-clinical tool compounds with a companion information resource” source here, so a public dataset of some drugs
- Redock scores: “This directory contains experiments in redocking all screened fragments into the entire ensemble of X-ray structures.” Taking fragments and re-docking them
repurposing_df = pd.read_csv('repurposing-screen/drugset-docked.csv')
redock_df = pd.read_csv('redock-fragments/all-screened-fragments-docked.csv')
SMILES strings, names, docking scores
repurposing_df.head()
| SMILES | TITLE | Hybrid2 | docked_fragment | Mpro-_dock | site | |
|---|---|---|---|---|---|---|
| 0 | C[C@@H](c1ccc-2c(c1)Cc3c2cccc3)C(=O)[O-] | CHEMBL2104122 | -11.519580 | x0749 | 0.509349 | active-covalent |
| 1 | C[C@]12CC[C@H]3[C@H]([C@@H]1CC[C@]2(C#C)O)CCC4... | CHEMBL1387 | -10.580162 | x0749 | 2.706928 | active-covalent |
| 2 | CC(C)(C)c1cc(cc(c1O)C(C)(C)C)/C=C\2/C(=O)NC(=[... | CHEMBL275835 | -10.557229 | x0107 | 1.801830 | active-noncovalent |
| 3 | C[C@]12CC[C@@H]3[C@H]4CCCCC4=CC[C@H]3[C@@H]1CC... | CHEMBL2104104 | -10.480992 | x0749 | 3.791700 | active-covalent |
| 4 | CC(=O)[C@]1(CC[C@@H]2[C@@]1(CCC3=C4CCC(=O)C=C4... | CHEMBL2104231 | -10.430775 | x0749 | 4.230903 | active-covalent |
Hybrid2 looks like a docking method provided via OpenEye. Mpro likely refers to COVID-19 main protease. I’m not entirely sure what the receptor for “Hybrid2” is, but there seem to be multiple “sites” or “fragments” for docking. There are lots of different fragments, but very few sites. For each site-fragment combination, multiple small molecules may have been tested.
repurposing_df['docked_fragment'].value_counts()
x0195 114
x0749 69
x0678 58
x0397 45
x0104 24
x0161 21
x1077 19
x0072 14
x0874 13
x0354 13
x0689 10
x1382 7
x0708 4
x0434 4
x1093 3
x1392 2
x0395 2
x1402 2
x0831 2
x0107 2
x1385 2
x1418 2
x0387 2
x0830 2
x1478 1
x0786 1
x1187 1
x0692 1
x0967 1
x0426 1
x0305 1
x0946 1
x1386 1
x0759 1
Name: docked_fragment, dtype: int64
repurposing_df['site'].value_counts()
active-noncovalent 338
active-covalent 107
dimer-interface 1
Name: site, dtype: int64
repurposing_df.groupby(["docked_fragment", "site"]).count()
| SMILES | TITLE | Hybrid2 | Mpro-_dock | ||
|---|---|---|---|---|---|
| docked_fragment | site | ||||
| x0072 | active-noncovalent | 14 | 14 | 14 | 14 |
| x0104 | active-noncovalent | 24 | 24 | 24 | 24 |
| x0107 | active-noncovalent | 2 | 2 | 2 | 2 |
| x0161 | active-noncovalent | 21 | 21 | 21 | 21 |
| x0195 | active-noncovalent | 114 | 114 | 114 | 114 |
| x0305 | active-noncovalent | 1 | 1 | 1 | 1 |
| x0354 | active-noncovalent | 13 | 13 | 13 | 13 |
| x0387 | active-noncovalent | 2 | 2 | 2 | 2 |
| x0395 | active-noncovalent | 2 | 2 | 2 | 2 |
| x0397 | active-noncovalent | 45 | 45 | 45 | 45 |
| x0426 | active-noncovalent | 1 | 1 | 1 | 1 |
| x0434 | active-noncovalent | 4 | 4 | 4 | 4 |
| x0678 | active-noncovalent | 58 | 58 | 58 | 58 |
| x0689 | active-covalent | 10 | 10 | 10 | 10 |
| x0692 | active-covalent | 1 | 1 | 1 | 1 |
| x0708 | active-covalent | 4 | 4 | 4 | 4 |
| x0749 | active-covalent | 69 | 69 | 69 | 69 |
| x0759 | active-covalent | 1 | 1 | 1 | 1 |
| x0786 | active-covalent | 1 | 1 | 1 | 1 |
| x0830 | active-covalent | 2 | 2 | 2 | 2 |
| x0831 | active-covalent | 2 | 2 | 2 | 2 |
| x0874 | active-noncovalent | 13 | 13 | 13 | 13 |
| x0946 | active-noncovalent | 1 | 1 | 1 | 1 |
| x0967 | active-noncovalent | 1 | 1 | 1 | 1 |
| x1077 | active-noncovalent | 19 | 19 | 19 | 19 |
| x1093 | active-noncovalent | 3 | 3 | 3 | 3 |
| x1187 | dimer-interface | 1 | 1 | 1 | 1 |
| x1382 | active-covalent | 7 | 7 | 7 | 7 |
| x1385 | active-covalent | 2 | 2 | 2 | 2 |
| x1386 | active-covalent | 1 | 1 | 1 | 1 |
| x1392 | active-covalent | 2 | 2 | 2 | 2 |
| x1402 | active-covalent | 2 | 2 | 2 | 2 |
| x1418 | active-covalent | 2 | 2 | 2 | 2 |
| x1478 | active-covalent | 1 | 1 | 1 | 1 |
Some molecules show up multiple times - why? Upon further investigation, this is mainly due to the molecule’s presence in multiple databases
repurposing_df.groupby(['SMILES']).count().sort_values("TITLE")
| TITLE | Hybrid2 | docked_fragment | Mpro-_dock | site | |
|---|---|---|---|---|---|
| SMILES | |||||
| B(CCCC)(O)O | 1 | 1 | 1 | 1 | 1 |
| CCCc1ccccc1N | 1 | 1 | 1 | 1 | 1 |
| CCCc1cc(=O)[nH]c(=S)[nH]1 | 1 | 1 | 1 | 1 | 1 |
| CCC[N@@H+]1CCO[C@H]2[C@H]1CCc3c2cc(cc3)O | 1 | 1 | 1 | 1 | 1 |
| CCC[N@@H+]1CCC[C@H]2[C@H]1Cc3c[nH]nc3C2 | 1 | 1 | 1 | 1 | 1 |
| ... | ... | ... | ... | ... | ... |
| C[C@]12CC[C@H]3[C@H]([C@@H]1CC[C@]2(C#C)O)CCC4=CC(=O)CC[C@H]34 | 2 | 2 | 2 | 2 | 2 |
| C[C@]12CC[C@H]3[C@H]([C@@H]1CCC2=O)CC(=C)C4=CC(=O)C=C[C@]34C | 2 | 2 | 2 | 2 | 2 |
| CC(C)C[C@@H](C1(CCC1)c2ccc(cc2)Cl)[NH+](C)C | 2 | 2 | 2 | 2 | 2 |
| CC[C@](/C=C/Cl)(C#C)O | 2 | 2 | 2 | 2 | 2 |
| CC[C@]12CC[C@H]3[C@H]([C@@H]1CC[C@@]2(C#C)O)CCC4=CC(=O)CC[C@H]34 | 2 | 2 | 2 | 2 | 2 |
432 rows × 5 columns
repurposing_df[repurposing_df['SMILES']=="CC[C@]12CC[C@H]3[C@H]([C@@H]1CC[C@@]2(C#C)O)CCC4=CC(=O)CC[C@H]34"]
| SMILES | TITLE | Hybrid2 | docked_fragment | Mpro-_dock | site | |
|---|---|---|---|---|---|---|
| 82 | CC[C@]12CC[C@H]3[C@H]([C@@H]1CC[C@@]2(C#C)O)CC... | CHEMBL2107797 | -9.002963 | x0749 | 2.616094 | active-covalent |
| 105 | CC[C@]12CC[C@H]3[C@H]([C@@H]1CC[C@@]2(C#C)O)CC... | EDRUG178 | -8.705896 | x0104 | 2.248707 | active-noncovalent |
There doesn’t seem to be a very good correlation between the two docking scores - if these are docking scores to different receptors, that would help explain things. It’s worth noting that we’re not seeing if the two numbers agree for each molecule, but if the trends persist (both scores go up for this molecule, but go down for this other molecule). The weak correlation suggests the trends do not persist between the two docking measures
repurposing_df[['Hybrid2', 'Mpro-_dock']].corr()
| Hybrid2 | Mpro-_dock | |
|---|---|---|
| Hybrid2 | 1.000000 | 0.581966 |
| Mpro-_dock | 0.581966 | 1.000000 |
Redocking dataframe: SMILES, names, data collection information, docking scores
redock_df.head()
| SMILES | TITLE | fragments | CompoundCode | Unnamed: 4 | covalent_warhead | MountingResult | DataCollectionOutcome | DataProcessingResolutionHigh | RefinementOutcome | Deposition_PDB_ID | Hybrid2 | docked_fragment | Mpro-x0500_dock | site | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | c1ccc(c(c1)NCc2ccn[nH]2)F | x0500 | x0500 | Z1545196403 | NaN | False | OK: No comment:No comment | success | 2.19 | 7 - Analysed & Rejected | NaN | -11.881923 | x0678 | -2.501554 | active-noncovalent |
| 1 | Cc1ccccc1OCC(=O)Nc2ncccn2 | x0415 | x0415 | Z53834613 | NaN | False | OK: No comment:No comment | success | 1.62 | 7 - Analysed & Rejected | NaN | -11.622278 | x0678 | NaN | active-noncovalent |
| 2 | Cc1csc(n1)CNC(=O)c2ccn[nH]2 | x0356 | x0356 | Z466628048 | NaN | False | OK: No comment:No comment | success | 3.25 | 7 - Analysed & Rejected | NaN | -11.435024 | x0678 | NaN | active-noncovalent |
| 3 | Cc1csc(n1)CNC(=O)c2ccn[nH]2 | x1113 | x1113 | Z466628048 | NaN | False | OK: No comment:No comment | success | 1.57 | 7 - Analysed & Rejected | NaN | -11.435024 | x0678 | NaN | active-noncovalent |
| 4 | c1cc(cnc1)NC(=O)CC2CCCCC2 | x0678 | x0678 | Z31792168 | NaN | False | Mounted_Clear | success | 1.83 | 6 - Deposited | 5R84 | -11.355046 | x0678 | NaN | active-noncovalent |
There don’t seem to be many Mpro docking scores in this dataset (only one molecule has a non-null Mpro docking score)
redock_df[redock_df['Mpro-x0500_dock'].isnull()].count()
SMILES 1452
TITLE 1452
fragments 1452
CompoundCode 1452
Unnamed: 4 0
covalent_warhead 1452
MountingResult 1452
DataCollectionOutcome 1452
DataProcessingResolutionHigh 1357
RefinementOutcome 1306
Deposition_PDB_ID 78
Hybrid2 1452
docked_fragment 1452
Mpro-x0500_dock 0
site 1452
dtype: int64
redock_df[~redock_df['Mpro-x0500_dock'].isnull()].count()
SMILES 1
TITLE 1
fragments 1
CompoundCode 1
Unnamed: 4 0
covalent_warhead 1
MountingResult 1
DataCollectionOutcome 1
DataProcessingResolutionHigh 1
RefinementOutcome 1
Deposition_PDB_ID 0
Hybrid2 1
docked_fragment 1
Mpro-x0500_dock 1
site 1
dtype: int64
Are there overlaps in the molecules in each of these datasets?
repurpose_redock = repurposing_df.merge(redock_df, on='SMILES', how='inner',suffixes=("_L", "_R"))
moonshot_redock = moonshot_df.merge(redock_df, on='SMILES', how='inner',suffixes=("_L", "_R"))
repurpose_redock
| SMILES | TITLE_L | Hybrid2_L | docked_fragment_L | Mpro-_dock | site_L | TITLE_R | fragments | CompoundCode | Unnamed: 4 | covalent_warhead | MountingResult | DataCollectionOutcome | DataProcessingResolutionHigh | RefinementOutcome | Deposition_PDB_ID | Hybrid2_R | docked_fragment_R | Mpro-x0500_dock | site_R | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Cc1cc(=O)n([nH]1)c2ccccc2 | CHEMBL290916 | -7.889587 | x0195 | -2.068452 | active-noncovalent | x0297 | x0297 | Z50145861 | NaN | False | OK: No comment:No comment | success | 1.98 | 7 - Analysed & Rejected | NaN | -7.889587 | x0195 | NaN | active-noncovalent |
| 1 | CC(C)Nc1ncccn1 | CHEMBL1740513 | -7.178702 | x0072 | -1.248482 | active-noncovalent | x0583 | x0583 | Z31190928 | NaN | False | OK: No comment:No comment | success | 3.08 | 7 - Analysed & Rejected | NaN | -7.293537 | x1093 | NaN | active-noncovalent |
| 2 | CC(C)Nc1ncccn1 | CHEMBL1740513 | -7.178702 | x0072 | -1.248482 | active-noncovalent | x1102 | x1102 | Z31190928 | NaN | False | OK: No comment:No comment | success | 1.46 | 7 - Analysed & Rejected | NaN | -7.293537 | x1093 | NaN | active-noncovalent |
| 3 | C[C@H](C(=O)[O-])O | CHEMBL1200559 | -5.675188 | x0397 | -0.179049 | active-noncovalent | x1035 | x1035 | Z1741982441 | NaN | False | OK: No comment:No comment | Failed - no diffraction | NaN | NaN | NaN | -6.505556 | x0397 | NaN | active-noncovalent |
| 4 | CC(=O)C(=O)[O-] | DB00119 | -5.448891 | x0689 | -0.494791 | active-covalent | x1037 | x1037 | Z1741977082 | NaN | False | OK: No comment:No comment | Failed - no diffraction | NaN | NaN | NaN | -5.448891 | x0689 | NaN | active-covalent |
| 5 | CCC(=O)[O-] | CHEMBL14021 | -5.374838 | x0397 | -0.555688 | active-noncovalent | x1029 | x1029 | Z955123616 | NaN | False | OK: No comment:No comment | success | 1.73 | 7 - Analysed & Rejected | NaN | -5.135675 | x0689 | NaN | active-covalent |
| 6 | C1CNCC[NH2+]1 | CHEMBL1412 | -5.079155 | x0354 | 1.716032 | active-noncovalent | x0996 | x0996 | Z1245537944 | NaN | False | OK: No comment:No comment | success | 1.96 | 7 - Analysed & Rejected | NaN | -4.675085 | x0354 | NaN | active-noncovalent |
We joined on SMILES string, and now we can compare the docking scores between the repurposing and redocking datasets.
Some Hybrid2 scores look quantitatively similar, but for those that don’t, the ranking is still there. Looking at the COVID-19 main protease (Mpro I believe?), the docking scores don’t follow similar rankings - docking scores aren’t transferable to different receptors (this might be a fairly obvious observation)
repurpose_redock[['SMILES', "TITLE_L", "TITLE_R", "Hybrid2_L", "Hybrid2_R", 'Mpro-_dock', 'Mpro-x0500_dock']]
| SMILES | TITLE_L | TITLE_R | Hybrid2_L | Hybrid2_R | Mpro-_dock | Mpro-x0500_dock | |
|---|---|---|---|---|---|---|---|
| 0 | Cc1cc(=O)n([nH]1)c2ccccc2 | CHEMBL290916 | x0297 | -7.889587 | -7.889587 | -2.068452 | NaN |
| 1 | CC(C)Nc1ncccn1 | CHEMBL1740513 | x0583 | -7.178702 | -7.293537 | -1.248482 | NaN |
| 2 | CC(C)Nc1ncccn1 | CHEMBL1740513 | x1102 | -7.178702 | -7.293537 | -1.248482 | NaN |
| 3 | C[C@H](C(=O)[O-])O | CHEMBL1200559 | x1035 | -5.675188 | -6.505556 | -0.179049 | NaN |
| 4 | CC(=O)C(=O)[O-] | DB00119 | x1037 | -5.448891 | -5.448891 | -0.494791 | NaN |
| 5 | CCC(=O)[O-] | CHEMBL14021 | x1029 | -5.374838 | -5.135675 | -0.555688 | NaN |
| 6 | C1CNCC[NH2+]1 | CHEMBL1412 | x0996 | -5.079155 | -4.675085 | 1.716032 | NaN |
Joining the moonshot submission and redocking datasets does not yield too many overlapping molecules
moonshot_redock
| SMILES | CID | creator | fragments_L | link | real_space | SCR | BB | extended_real_space | in_molport_or_mcule | in_ultimate_mcule | in_emolecules | covalent_frag | covalent_warhead_L | acrylamide | acrylamide_adduct | chloroacetamide | chloroacetamide_adduct | vinylsulfonamide | vinylsulfonamide_adduct | nitrile | nitrile_adduct | MW | cLogP | HBD | HBA | TPSA | num_criterion_violations | BMS | Dundee | Glaxo | Inpharmatica | LINT | MLSMR | PAINS | SureChEMBL | PostEra | ORDERED | MADE | ASSAYED | TITLE | fragments_R | CompoundCode | Unnamed: 4 | covalent_warhead_R | MountingResult | DataCollectionOutcome | DataProcessingResolutionHigh | RefinementOutcome | Deposition_PDB_ID | Hybrid2 | docked_fragment | Mpro-x0500_dock | site | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CC(C)Nc1cccnc1 | MAK-UNK-2c1752f0-4 | Maksym Voznyy | x1093 | https://covid.postera.ai/covid/submissions/MAK... | FALSE | Z2574930241 | EN300-56005 | FALSE | False | False | False | False | False | False | False | False | False | False | False | False | False | 136.198 | 1.9019 | 1 | 2 | 24.92 | 0 | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | False | False | False | x1098 | x1098 | Z1259341037 | NaN | False | OK: No comment:No comment | success | 1.66 | 7 - Analysed & Rejected | NaN | -7.474369 | x0678 | NaN | active-noncovalent |
| 1 | CC(C)Nc1cccnc1 | MAK-UNK-2c1752f0-4 | Maksym Voznyy | x1093 | https://covid.postera.ai/covid/submissions/MAK... | FALSE | Z2574930241 | EN300-56005 | FALSE | False | False | False | False | False | False | False | False | False | False | False | False | False | 136.198 | 1.9019 | 1 | 2 | 24.92 | 0 | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | False | False | False | x0572 | x0572 | Z1259341037 | NaN | False | OK: No comment:No comment | success | 2.98 | 7 - Analysed & Rejected | NaN | -7.474369 | x0678 | NaN | active-noncovalent |
| 2 | CCS(=O)(=O)Nc1ccccc1F | MAK-UNK-2c1752f0-5 | Maksym Voznyy | x1093 | https://covid.postera.ai/covid/submissions/MAK... | FALSE | Z53825177 | EN300-116204 | FALSE | False | True | False | False | False | False | False | False | False | False | False | False | False | 203.238 | 1.5873 | 1 | 2 | 46.17 | 0 | PASS | PASS | PASS | PASS | PASS | Hetero_hetero | PASS | PASS | PASS | False | False | False | x0247 | x0247 | Z53825177 | NaN | False | OK: No comment:No comment | success | 1.83 | 7 - Analysed & Rejected | NaN | -7.413380 | x0678 | NaN | active-noncovalent |
Comparing other databases
CHEMBL, DrugBank, and “EDrug”(?) look to be the 3 prefixes in the “TITLE” column
from chembl_webresource_client.new_client import new_client
molecule = new_client.molecule
res = molecule.search('CHEMBL1387')
res_df = pd.DataFrame.from_dict(res)
res_df.columns
Index(['atc_classifications', 'availability_type', 'biotherapeutic',
'black_box_warning', 'chebi_par_id', 'chirality', 'cross_references',
'dosed_ingredient', 'first_approval', 'first_in_class', 'helm_notation',
'indication_class', 'inorganic_flag', 'max_phase', 'molecule_chembl_id',
'molecule_hierarchy', 'molecule_properties', 'molecule_structures',
'molecule_synonyms', 'molecule_type', 'natural_product', 'oral',
'parenteral', 'polymer_flag', 'pref_name', 'prodrug', 'score',
'structure_type', 'therapeutic_flag', 'topical', 'usan_stem',
'usan_stem_definition', 'usan_substem', 'usan_year', 'withdrawn_class',
'withdrawn_country', 'withdrawn_flag', 'withdrawn_reason',
'withdrawn_year'],
dtype='object')
res_df[['chirality', 'molecule_properties', 'molecule_structures', 'score']]
| chirality | molecule_properties | molecule_structures | score | |
|---|---|---|---|---|
| 0 | 1 | {'alogp': '3.64', 'aromatic_rings': 0, 'cx_log... | {'canonical_smiles': 'C#C[C@]1(O)CC[C@H]2[C@@H... | 17.0 |
res_df[['molecule_properties']].values[0]
array([{'alogp': '3.64', 'aromatic_rings': 0, 'cx_logd': '2.81', 'cx_logp': '2.81', 'cx_most_apka': None, 'cx_most_bpka': None, 'full_molformula': 'C20H26O2', 'full_mwt': '298.43', 'hba': 2, 'hba_lipinski': 2, 'hbd': 1, 'hbd_lipinski': 1, 'heavy_atoms': 22, 'molecular_species': None, 'mw_freebase': '298.43', 'mw_monoisotopic': '298.1933', 'num_lipinski_ro5_violations': 0, 'num_ro5_violations': 0, 'psa': '37.30', 'qed_weighted': '0.55', 'ro3_pass': 'N', 'rtb': 0}],
dtype=object)
res_df['molecule_properties'].apply(pd.Series)
| alogp | aromatic_rings | cx_logd | cx_logp | cx_most_apka | cx_most_bpka | full_molformula | full_mwt | hba | hba_lipinski | hbd | hbd_lipinski | heavy_atoms | molecular_species | mw_freebase | mw_monoisotopic | num_lipinski_ro5_violations | num_ro5_violations | psa | qed_weighted | ro3_pass | rtb | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 3.64 | 0 | 2.81 | 2.81 | None | None | C20H26O2 | 298.43 | 2 | 2 | 1 | 1 | 22 | None | 298.43 | 298.1933 | 0 | 0 | 37.30 | 0.55 | N | 0 |
all_results = [molecule.search(a) for a in repurposing_df['TITLE']]
Here’s a big Python function tangent.
For each chembl molecule, we’ve searched for it within the chembl, returning us a list (of length 1) containing a dictionary of properties.
All molecules have been compiled into a list, so we have a list of lists of dicionatires.
For sanity, we can use a Python filter to only retain the non-None results.
We can chain that with a Python map function to parse the first item from each molecule’s list. Recall, each molecule was a list with just one element, a dictionary. We can boil this down to only returning the dictionary (eliminating the list wrapper).
For validation, I’ve called next to look at the results
filtered = map(lambda x: x[0], filter(lambda x: x is not None, all_results))
next(filtered)
{'atc_classifications': [],
'availability_type': -1,
'biotherapeutic': None,
'black_box_warning': 0,
'chebi_par_id': None,
'chirality': 0,
'cross_references': [],
'dosed_ingredient': False,
'first_approval': None,
'first_in_class': 0,
'helm_notation': None,
'indication_class': 'Anti-Inflammatory',
'inorganic_flag': 0,
'max_phase': 0,
'molecule_chembl_id': 'CHEMBL2104122',
'molecule_hierarchy': {'molecule_chembl_id': 'CHEMBL2104122',
'parent_chembl_id': 'CHEMBL2104122'},
'molecule_properties': {'alogp': '3.45',
'aromatic_rings': 2,
'cx_logd': '1.26',
'cx_logp': '3.92',
'cx_most_apka': '4.68',
'cx_most_bpka': None,
'full_molformula': 'C16H14O2',
'full_mwt': '238.29',
'hba': 1,
'hba_lipinski': 2,
'hbd': 1,
'hbd_lipinski': 1,
'heavy_atoms': 18,
'molecular_species': 'ACID',
'mw_freebase': '238.29',
'mw_monoisotopic': '238.0994',
'num_lipinski_ro5_violations': 0,
'num_ro5_violations': 0,
'psa': '37.30',
'qed_weighted': '0.74',
'ro3_pass': 'N',
'rtb': 2},
'molecule_structures': {'canonical_smiles': 'CC(C(=O)O)c1ccc2c(c1)Cc1ccccc1-2',
'molfile': '\n RDKit 2D\n\n 18 20 0 0 0 0 0 0 0 0999 V2000\n -0.5375 0.0250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n -0.5375 1.1083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n -2.4458 1.1083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n -2.4458 0.0250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 1.3625 0.0250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n -1.4875 -0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 0.4125 -0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 3.3292 0.0250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 0.4125 1.6500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 2.3417 -0.5292 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 1.3625 1.1083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 3.3500 1.1958 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0\n 4.2167 -0.6292 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0\n -3.3958 1.6500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n -3.3958 -0.5125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 2.3417 -1.6417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n -4.3458 1.1083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n -4.3458 0.0250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 2 1 2 0\n 3 2 1 0\n 4 6 1 0\n 5 7 2 0\n 6 1 1 0\n 7 1 1 0\n 8 10 1 0\n 9 2 1 0\n 10 5 1 0\n 11 5 1 0\n 12 8 2 0\n 13 8 1 0\n 14 3 1 0\n 15 4 1 0\n 16 10 1 0\n 17 14 2 0\n 18 15 2 0\n 3 4 2 0\n 9 11 2 0\n 17 18 1 0\nM END\n\n> <chembl_id>\nCHEMBL2104122\n\n> <chembl_pref_name>\nCICLOPROFEN\n\n',
'standard_inchi': 'InChI=1S/C16H14O2/c1-10(16(17)18)11-6-7-15-13(8-11)9-12-4-2-3-5-14(12)15/h2-8,10H,9H2,1H3,(H,17,18)',
'standard_inchi_key': 'LRXFKKPEBXIPMW-UHFFFAOYSA-N'},
'molecule_synonyms': [{'molecule_synonym': 'Cicloprofen',
'syn_type': 'BAN',
'synonyms': 'CICLOPROFEN'},
{'molecule_synonym': 'Cicloprofen',
'syn_type': 'INN',
'synonyms': 'CICLOPROFEN'},
{'molecule_synonym': 'Cicloprofen',
'syn_type': 'USAN',
'synonyms': 'CICLOPROFEN'},
{'molecule_synonym': 'SQ-20824',
'syn_type': 'RESEARCH_CODE',
'synonyms': 'SQ 20824'}],
'molecule_type': 'Small molecule',
'natural_product': 0,
'oral': False,
'parenteral': False,
'polymer_flag': False,
'pref_name': 'CICLOPROFEN',
'prodrug': 0,
'score': 16.0,
'structure_type': 'MOL',
'therapeutic_flag': False,
'topical': False,
'usan_stem': '-profen',
'usan_stem_definition': 'anti-inflammatory/analgesic agents (ibuprofen type)',
'usan_substem': '-profen',
'usan_year': 1974,
'withdrawn_class': None,
'withdrawn_country': None,
'withdrawn_flag': False,
'withdrawn_reason': None,
'withdrawn_year': None}
For now, I’m only really interested in the molecule_properties dictionary
filtered = [a[0]['molecule_properties'] for a in all_results if len(a) > 0]
chembl_df = pd.DataFrame(filtered)
chembl_df['TITLE'] = repurposing_df['TITLE']
Molecular properties contained in the chembl database
Here are the definitions I can dig up
- alogp: (lipophilicity) partition coefficient
- aromatic_rings: number of aromatic rings
- cx_logd: distribution coefficient taking into account ionized and non-ionized forms
- cx_most_apka: acidic pka
- cx_most_bpka: basic pka
- full_mwt: molecular weight (and also free base and monoisotopic masses)
- hba: hydrogen bond acceptors (and hba_lipinski for lipinski definitiosn)
- hbd: hydrogen bond donors (and hbd_lipinski)
- heavy_atoms: number of heavy atoms
- num_lipinski_ro5_violations: how many times this molecule violated Lipinski’s rule of five
- num_ro5_violations: not sure, seems similar to lipinski rule of 5
- psa: protein sequence alignment
- qed_weighted: “quantitative estimate of druglikeness” (ranges between 0 and 1, with 1 being more favorable). This is based on a quantitatve mean of drugability functions
- ro3_pass: rule of three
- rtb: number of rotatable bonds
chembl_df.head()
| alogp | aromatic_rings | cx_logd | cx_logp | cx_most_apka | cx_most_bpka | full_molformula | full_mwt | hba | hba_lipinski | hbd | hbd_lipinski | heavy_atoms | molecular_species | mw_freebase | mw_monoisotopic | num_lipinski_ro5_violations | num_ro5_violations | psa | qed_weighted | ro3_pass | rtb | TITLE | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 3.45 | 2.0 | 1.26 | 3.92 | 4.68 | None | C16H14O2 | 238.29 | 1.0 | 2.0 | 1.0 | 1.0 | 18.0 | ACID | 238.29 | 238.0994 | 0.0 | 0.0 | 37.30 | 0.74 | N | 2.0 | CHEMBL2104122 |
| 1 | 3.64 | 0.0 | 2.81 | 2.81 | None | None | C20H26O2 | 298.43 | 2.0 | 2.0 | 1.0 | 1.0 | 22.0 | None | 298.43 | 298.1933 | 0.0 | 0.0 | 37.30 | 0.55 | N | 0.0 | CHEMBL1387 |
| 2 | 3.92 | 1.0 | 4.25 | 4.25 | 10.15 | 2.86 | C18H24N2O2S | 332.47 | 4.0 | 4.0 | 2.0 | 3.0 | 23.0 | NEUTRAL | 332.47 | 332.1558 | 0.0 | 0.0 | 75.68 | 0.76 | N | 1.0 | CHEMBL275835 |
| 3 | 4.31 | 0.0 | 4.04 | 4.04 | None | None | C20H28O | 284.44 | 1.0 | 1.0 | 1.0 | 1.0 | 21.0 | None | 284.44 | 284.2140 | 0.0 | 0.0 | 20.23 | 0.52 | N | 0.0 | CHEMBL2104104 |
| 4 | 4.79 | 0.0 | 3.96 | 3.96 | None | None | C21H28O2 | 312.45 | 2.0 | 2.0 | 0.0 | 0.0 | 23.0 | None | 312.45 | 312.2089 | 0.0 | 0.0 | 34.14 | 0.70 | N | 1.0 | CHEMBL2104231 |
chembl_df.columns
Index(['alogp', 'aromatic_rings', 'cx_logd', 'cx_logp', 'cx_most_apka',
'cx_most_bpka', 'full_molformula', 'full_mwt', 'hba', 'hba_lipinski',
'hbd', 'hbd_lipinski', 'heavy_atoms', 'molecular_species',
'mw_freebase', 'mw_monoisotopic', 'num_lipinski_ro5_violations',
'num_ro5_violations', 'psa', 'qed_weighted', 'ro3_pass', 'rtb',
'TITLE'],
dtype='object')
chembl_df.corr()
| aromatic_rings | hba | hba_lipinski | hbd | hbd_lipinski | heavy_atoms | num_lipinski_ro5_violations | num_ro5_violations | rtb | |
|---|---|---|---|---|---|---|---|---|---|
| aromatic_rings | 1.000000 | 0.192569 | 0.178507 | 0.014928 | 0.036106 | 0.249022 | 0.031094 | 0.031094 | 0.229124 |
| hba | 0.192569 | 1.000000 | 0.868859 | 0.084553 | 0.054409 | 0.451560 | -0.047705 | -0.047705 | -0.023690 |
| hba_lipinski | 0.178507 | 0.868859 | 1.000000 | 0.348600 | 0.294276 | 0.295864 | -0.070783 | -0.070783 | 0.021812 |
| hbd | 0.014928 | 0.084553 | 0.348600 | 1.000000 | 0.935710 | -0.172866 | -0.060462 | -0.060462 | 0.040505 |
| hbd_lipinski | 0.036106 | 0.054409 | 0.294276 | 0.935710 | 1.000000 | -0.211899 | -0.085660 | -0.085660 | 0.084225 |
| heavy_atoms | 0.249022 | 0.451560 | 0.295864 | -0.172866 | -0.211899 | 1.000000 | 0.397240 | 0.397240 | 0.259011 |
| num_lipinski_ro5_violations | 0.031094 | -0.047705 | -0.070783 | -0.060462 | -0.085660 | 0.397240 | 1.000000 | 1.000000 | 0.345308 |
| num_ro5_violations | 0.031094 | -0.047705 | -0.070783 | -0.060462 | -0.085660 | 0.397240 | 1.000000 | 1.000000 | 0.345308 |
| rtb | 0.229124 | -0.023690 | 0.021812 | 0.040505 | 0.084225 | 0.259011 | 0.345308 | 0.345308 | 1.000000 |
At a glance, no definite linear correlations among this crowd besides pKas, partition coefficients, mwt/hba
corr_df = chembl_df.corr()
cols = chembl_df.columns
fig, ax = plt.subplots(1,1, figsize=(8,6), dpi=100)
ax.imshow(chembl_df.corr(), cmap='RdBu')
ax.set_xticklabels(['']+cols)
ax.tick_params(axis='x', rotation=90)
ax.set_yticklabels(cols)
for i, (rowname, row) in enumerate(corr_df.iterrows()):
for j, (key, val) in enumerate(row.iteritems()):
ax.annotate(f"{val:0.2f}", xy=(i,j), xytext=(-10, -5), textcoords="offset points")
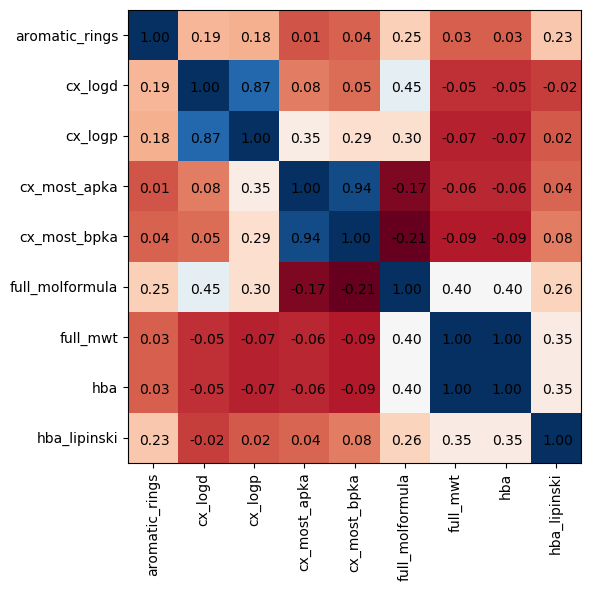
Maybe there are higher-order correlations and relationship more appropriate for clustering and decomposition
cols = ['aromatic_rings', 'cx_logp', 'full_mwt', 'hba']
cleaned = (chembl_df[~chembl_df[cols]
.isnull()
.all(axis='columns', skipna=False)][cols]
.astype('float')
.fillna(0, axis='columns'))
from sklearn import preprocessing
normalized = preprocessing.scale(cleaned)
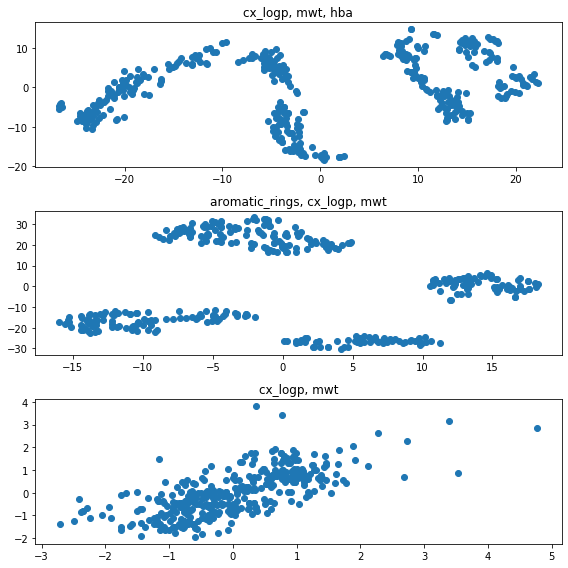
Appears to be maybe 4 clusters of these compounds examined by the covid-moonshot group
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
tsne_analysis = TSNE(n_components=2)
output = tsne_analysis.fit_transform(normalized)
fig,ax = plt.subplots(1,1)
ax.scatter(output[:,0], output[:,1])
ax.set_title("Aromatic rings, cx_logp, mwt, hba")
Text(0.5, 1.0, 'Aromatic rings, cx_logp, mwt, hba')

By taking turns leaving out some features, it looks like leaving out aromatic rings or hydrogen bond acceptors will diminish the cluster distinction.
Aromatic rings are huge and bulky components to small molecules, it makes sense that a chunk of the behavior corresponds to the aromatic rings. Similarly, hydrogen bond acceptors (heavy molecules) also induce van der Waals and electrostatics influences on small molecules. Left with only weight and partition coefficient, there’s mainly a continous behavior
def clean_df(cols):
cleaned = (chembl_df[~chembl_df[cols]
.isnull()
.all(axis='columns', skipna=False)][cols]
.astype('float')
.fillna(0, axis='columns'))
normalized = preprocessing.scale(cleaned)
return normalized
cols = ['cx_logp', 'full_mwt', 'hba']
normalized = clean_df(cols)
tsne_analysis = TSNE(n_components=2)
output = tsne_analysis.fit_transform(normalized)
fig,ax = plt.subplots(3,1, figsize=(8,8))
ax[0].scatter(output[:,0], output[:,1])
ax[0].set_title("cx_logp, mwt, hba")
cols = ['cx_logp', 'full_mwt', 'aromatic_rings']
normalized = clean_df(cols)
tsne_analysis = TSNE(n_components=2)
output = tsne_analysis.fit_transform(normalized)
ax[1].scatter(output[:,0], output[:,1])
ax[1].set_title("aromatic_rings, cx_logp, mwt")
cols = ['cx_logp', 'full_mwt']
normalized = clean_df(cols)
ax[2].scatter(normalized[:,0], normalized[:,1])
ax[2].set_title("cx_logp, mwt")
fig.tight_layout()
DrugBank
I found someone had already downloaded the database. I may double-over these dataframes, but query the drugbank dataset rather than chembl
Some docking data
We have some smiles strings, molecular properties, docking scores, and information about the docking fragments
moonshot = pd.read_csv('moonshot-submissions/covid_submissions_all_info-docked-overlap.csv')
moonshot
| SMILES | TITLE | creator | fragments | link | real_space | SCR | BB | extended_real_space | in_molport_or_mcule | in_ultimate_mcule | in_emolecules | covalent_frag | covalent_warhead | acrylamide | acrylamide_adduct | chloroacetamide | chloroacetamide_adduct | vinylsulfonamide | vinylsulfonamide_adduct | nitrile | nitrile_adduct | MW | cLogP | HBD | HBA | TPSA | num_criterion_violations | BMS | Dundee | Glaxo | Inpharmatica | LINT | MLSMR | PAINS | SureChEMBL | PostEra | ORDERED | MADE | ASSAYED | Hybrid2 | docked_fragment | Mpro-x1418_dock | site | number_of_overlapping_fragments | overlapping_fragments | overlap_score | volume | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | c1ccc(cc1)n2c3cc(c(cc3c(=O)c(c2[O-])c4cccnc4)F)Cl | MAK-UNK-9e4a73aa-2 | Maksym Voznyy | x1418 | https://covid.postera.ai/covid/submissions/MAK... | FALSE | FALSE | FALSE | FALSE | False | False | False | True | False | False | False | False | False | False | False | False | False | 366.779 | 4.51890 | 0 | 3 | 50.27 | 0 | PASS | beta-keto/anhydride | PASS | PASS | PASS | Ketone, Dye 11 | PASS | PASS | PASS | False | False | False | -11.881256 | x1418 | 1.206534 | active-covalent | 3 | x0434,x0678,x0830 | 3.208124 | 271.986084 |
| 1 | Cc1ccncc1n2c(=O)ccc3c2CCCN3CC(=[NH2+])N | KIM-UNI-60f168f5-7 | Kim Tai Tran, University of Copenhagen | x0107,x0991 | https://covid.postera.ai/covid/submissions/KIM... | FALSE | FALSE | FALSE | FALSE | False | False | False | False | False | False | False | False | False | False | False | False | False | 297.362 | 1.22949 | 2 | 5 | 88.00 | 0 | PASS | imine, imine | PASS | PASS | acyclic C=N-H | Imine 3 | PASS | PASS | PASS | False | False | False | -11.654112 | x0107 | NaN | active-noncovalent | 3 | x0107,x1412,x1392 | 4.753475 | 232.815506 |
| 2 | c1ccc(cc1)n2c3cc(c(cc3c(=O)n(c2=O)c4cnccn4)F)Cl | MAK-UNK-9e4a73aa-14 | Maksym Voznyy | x1418 | https://covid.postera.ai/covid/submissions/MAK... | FALSE | FALSE | FALSE | FALSE | False | False | False | True | False | False | False | False | False | False | False | False | False | 368.755 | 2.72410 | 0 | 6 | 69.78 | 0 | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | False | False | False | -10.460650 | x0678 | 2.716276 | active-noncovalent | 3 | x0678,x1412,x1392 | 5.520980 | 266.688721 |
| 3 | Cc1ccncc1N(C=C)[C@H]([C@@H](C)[C@@H]2CN=Cc3c2c... | AUS-WAB-916db9c0-1 | Austin D. Chivington, Wabash College | x0107,x1077,x1374 | https://covid.postera.ai/covid/submissions/AUS... | FALSE | FALSE | FALSE | FALSE | False | False | False | True | False | False | False | False | False | False | False | False | False | 351.450 | 3.51932 | 1 | 5 | 57.95 | 0 | non_ring_acetal | het-C-het not in ring | PASS | Filter10_Terminal_vinyl | PASS | PASS | PASS | PASS | PASS | False | False | False | -9.516450 | x0678 | NaN | active-noncovalent | 3 | x0434,x0831,x0678 | 3.446572 | 284.195312 |
| 4 | c1ccc2c(c1)ncc(n2)/C=C/C(=O)c3cccc(c3)O | DRV-DNY-ae159ed1-12 | Dr. Vidya Desai, Dnyanprassarak Mandals Colleg... | x1249 | https://covid.postera.ai/covid/submissions/DRV... | FALSE | FALSE | FALSE | FALSE | False | False | False | False | False | False | False | False | False | False | False | False | False | 276.295 | 3.23150 | 1 | 4 | 63.08 | 0 | PASS | PASS | PASS | Filter44_michael_acceptor2 | PASS | Ketone, Dye 9, vinyl michael acceptor1 | PASS | PASS | PASS | False | False | False | -9.243208 | x0678 | NaN | active-noncovalent | 3 | x0434,x0678,x0830 | 2.865147 | 220.275421 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 4630 | C[C@H]([C@@H](C(=O)N[C@H](Cc1ccccc1)C(=O)N[C@@... | PAU-UNI-6d15a9f5-4 | paul brear, University of cambridge | x1086 | https://covid.postera.ai/covid/submissions/PAU... | FALSE | FALSE | FALSE | FALSE | False | False | False | False | False | False | False | False | False | False | False | False | False | 714.821 | -0.91270 | 8 | 11 | 256.10 | 4 | PASS | PASS | PASS | PASS | PASS | Long aliphatic chain, Dipeptide | PASS | PASS | PASS | False | False | False | 3.175111 | x0305 | NaN | active-noncovalent | 0 | NaN | 5.297134 | 548.583191 |
| 4631 | c1cc2cc(c(cc2c(c1)S(=O)(=O)N3CC[NH+](CC3)Cc4cc... | MAK-UNK-e05327b2-2 | Maksym Voznyy | x1402 | https://covid.postera.ai/covid/submissions/MAK... | FALSE | FALSE | FALSE | FALSE | False | False | False | True | True | False | False | True | False | False | False | False | False | 837.964 | 6.63190 | 0 | 9 | 98.31 | 2 | PASS | PASS | PASS | PASS | PASS | Hetero_hetero | PASS | PASS | PASS | False | False | False | 3.561681 | x1392 | NaN | active-covalent | 0 | NaN | 3.297014 | 591.877563 |
| 4632 | Cc1cccc(c1)C[NH+]2CCN(CC2)C(=O)c3ccc(cc3)C#Cc4... | MAK-UNK-e4a48a85-16 | Maksym Voznyy | x0387,x0692 | https://covid.postera.ai/covid/submissions/MAK... | FALSE | FALSE | FALSE | FALSE | False | False | False | True | False | False | False | False | False | False | False | False | False | 574.794 | 6.18892 | 0 | 5 | 39.68 | 2 | PASS | triple bond | PASS | PASS | PASS | PASS | PASS | PASS | PASS | False | False | False | 4.056698 | x0978 | NaN | active-covalent | 0 | NaN | 4.360606 | 470.944824 |
| 4633 | c1cc2cc(c(cc2c(c1)S(=O)(=O)N3CC[NH+](CC3)Cc4cc... | MAK-UNK-e05327b2-6 | Maksym Voznyy | x1402 | https://covid.postera.ai/covid/submissions/MAK... | FALSE | FALSE | FALSE | FALSE | False | False | False | True | True | False | False | True | False | False | False | False | False | 990.183 | 5.19160 | 0 | 12 | 138.93 | 3 | alpha_halo_heteroatom, secondary_halide_sulfate | PASS | PASS | PASS | PASS | Hetero_hetero | PASS | Dithiomethylene_acetal | Alkyl Halide | False | False | False | 4.242827 | x0731 | NaN | active-covalent | 0 | NaN | 4.193186 | 694.333069 |
| 4634 | Cc1cccc(c1)C[NH+]2CCN(CC2)c3cc(c(c(c3)Cl)c4cc5... | MAK-UNK-e4a48a85-15 | Maksym Voznyy | x0387,x0692 | https://covid.postera.ai/covid/submissions/MAK... | FALSE | FALSE | FALSE | FALSE | False | False | False | True | False | False | False | False | False | False | False | False | False | 659.687 | 7.36362 | 1 | 7 | 68.36 | 2 | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | False | False | False | 5.966927 | x0705 | NaN | active-covalent | 0 | NaN | 1.473711 | 503.583801 |
4635 rows × 48 columns
moonshot.head(5)
| SMILES | TITLE | creator | fragments | link | real_space | SCR | BB | extended_real_space | in_molport_or_mcule | in_ultimate_mcule | in_emolecules | covalent_frag | covalent_warhead | acrylamide | acrylamide_adduct | chloroacetamide | chloroacetamide_adduct | vinylsulfonamide | vinylsulfonamide_adduct | nitrile | nitrile_adduct | MW | cLogP | HBD | HBA | TPSA | num_criterion_violations | BMS | Dundee | Glaxo | Inpharmatica | LINT | MLSMR | PAINS | SureChEMBL | PostEra | ORDERED | MADE | ASSAYED | Hybrid2 | docked_fragment | Mpro-x1418_dock | site | number_of_overlapping_fragments | overlapping_fragments | overlap_score | volume | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | c1ccc(cc1)n2c3cc(c(cc3c(=O)c(c2[O-])c4cccnc4)F)Cl | MAK-UNK-9e4a73aa-2 | Maksym Voznyy | x1418 | https://covid.postera.ai/covid/submissions/MAK... | FALSE | FALSE | FALSE | FALSE | False | False | False | True | False | False | False | False | False | False | False | False | False | 366.779 | 4.51890 | 0 | 3 | 50.27 | 0 | PASS | beta-keto/anhydride | PASS | PASS | PASS | Ketone, Dye 11 | PASS | PASS | PASS | False | False | False | -11.881256 | x1418 | 1.206534 | active-covalent | 3 | x0434,x0678,x0830 | 3.208124 | 271.986084 |
| 1 | Cc1ccncc1n2c(=O)ccc3c2CCCN3CC(=[NH2+])N | KIM-UNI-60f168f5-7 | Kim Tai Tran, University of Copenhagen | x0107,x0991 | https://covid.postera.ai/covid/submissions/KIM... | FALSE | FALSE | FALSE | FALSE | False | False | False | False | False | False | False | False | False | False | False | False | False | 297.362 | 1.22949 | 2 | 5 | 88.00 | 0 | PASS | imine, imine | PASS | PASS | acyclic C=N-H | Imine 3 | PASS | PASS | PASS | False | False | False | -11.654112 | x0107 | NaN | active-noncovalent | 3 | x0107,x1412,x1392 | 4.753475 | 232.815506 |
| 2 | c1ccc(cc1)n2c3cc(c(cc3c(=O)n(c2=O)c4cnccn4)F)Cl | MAK-UNK-9e4a73aa-14 | Maksym Voznyy | x1418 | https://covid.postera.ai/covid/submissions/MAK... | FALSE | FALSE | FALSE | FALSE | False | False | False | True | False | False | False | False | False | False | False | False | False | 368.755 | 2.72410 | 0 | 6 | 69.78 | 0 | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | PASS | False | False | False | -10.460650 | x0678 | 2.716276 | active-noncovalent | 3 | x0678,x1412,x1392 | 5.520980 | 266.688721 |
| 3 | Cc1ccncc1N(C=C)[C@H]([C@@H](C)[C@@H]2CN=Cc3c2c... | AUS-WAB-916db9c0-1 | Austin D. Chivington, Wabash College | x0107,x1077,x1374 | https://covid.postera.ai/covid/submissions/AUS... | FALSE | FALSE | FALSE | FALSE | False | False | False | True | False | False | False | False | False | False | False | False | False | 351.450 | 3.51932 | 1 | 5 | 57.95 | 0 | non_ring_acetal | het-C-het not in ring | PASS | Filter10_Terminal_vinyl | PASS | PASS | PASS | PASS | PASS | False | False | False | -9.516450 | x0678 | NaN | active-noncovalent | 3 | x0434,x0831,x0678 | 3.446572 | 284.195312 |
| 4 | c1ccc2c(c1)ncc(n2)/C=C/C(=O)c3cccc(c3)O | DRV-DNY-ae159ed1-12 | Dr. Vidya Desai, Dnyanprassarak Mandals Colleg... | x1249 | https://covid.postera.ai/covid/submissions/DRV... | FALSE | FALSE | FALSE | FALSE | False | False | False | False | False | False | False | False | False | False | False | False | False | 276.295 | 3.23150 | 1 | 4 | 63.08 | 0 | PASS | PASS | PASS | Filter44_michael_acceptor2 | PASS | Ketone, Dye 9, vinyl michael acceptor1 | PASS | PASS | PASS | False | False | False | -9.243208 | x0678 | NaN | active-noncovalent | 3 | x0434,x0678,x0830 | 2.865147 | 220.275421 |
moonshot['Mpro-x1418_dock'].isnull().sum() # Lots of missing Mpro dock scores
4586
While there are a lot of different fragments to which the small molecule can bind, there are two “classes”, active-covalent and active-noncovalent (possibly referring to sites that covalently bond?)
This presents a way to logically bisect the data based on some fundamental chemistry of the binding pocket.
moonshot['docked_fragment'].value_counts()
x0678 940
x0749 771
x0104 347
x0831 283
x0830 281
x0195 269
x0161 252
x0107 201
x0072 172
x1077 127
x1392 107
x1093 107
x0434 105
x0874 81
x1385 69
x1418 58
x1334 50
x0967 46
x0397 42
x0946 38
x0692 37
x0759 37
x1386 35
x0395 29
x0305 24
x1311 16
x0708 13
x0774 12
x1380 10
x1412 7
x1374 7
x1348 6
x0770 5
x1249 5
x0387 5
x0736 4
x0705 4
x1358 3
x0426 3
x1375 3
x0734 3
x0540 3
x0354 3
x1382 3
x0755 1
x1458 1
x0689 1
x0769 1
x0981 1
x0978 1
x0731 1
x1493 1
x0771 1
x1478 1
x1384 1
x1351 1
Name: docked_fragment, dtype: int64
moonshot['site'].value_counts()
active-noncovalent 2799
active-covalent 1836
Name: site, dtype: int64
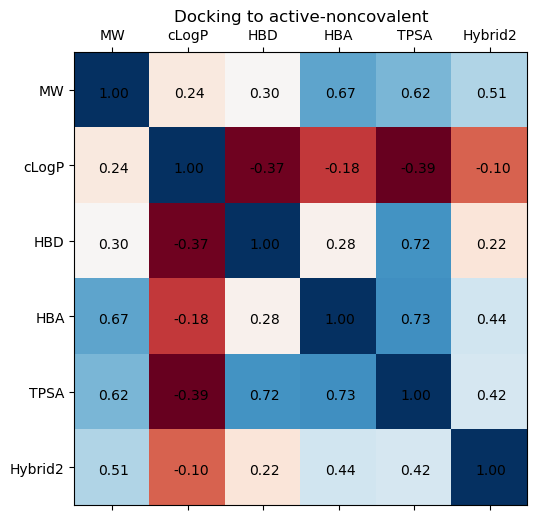
We can examine the same correlations, but now for each type of site, and look at the hybrid docking score correlations.
The biggest trend differences appear with the partition coefficient and number of hydrogen bond donors, but still the correlations are extremely weak
site_type = 'active-noncovalent'
fig, ax = plt.subplots(1,1, figsize=(8,6), dpi=100)
cols = ['MW', 'cLogP', 'HBD', 'HBA', 'TPSA', 'Hybrid2']
ax.matshow(moonshot[moonshot['site']==site_type][cols].corr(), cmap='RdBu')
ax.set_xticks([i for i,_ in enumerate(cols)])
ax.set_xticklabels(cols)
ax.set_yticks([i for i,_ in enumerate(cols)])
ax.set_yticklabels(cols)
for i, (rowname, row) in enumerate(moonshot[moonshot['site']==site_type][cols].corr().iterrows()):
for j, (key, val) in enumerate(row.iteritems()):
ax.annotate(f"{val:0.2f}", xy=(i,j), xytext=(-10, -5), textcoords="offset points")
ax.set_title(f"Docking to {site_type}")
Text(0.5, 1.05, 'Docking to active-noncovalent')
site_type = 'active-covalent'
fig, ax = plt.subplots(1,1, figsize=(8,6), dpi=100)
cols = ['MW', 'cLogP', 'HBD', 'HBA', 'TPSA', 'Hybrid2']
ax.matshow(moonshot[moonshot['site']==site_type][cols].corr(), cmap='RdBu')
ax.set_xticks([i for i,_ in enumerate(cols)])
ax.set_xticklabels(cols)
ax.set_yticks([i for i,_ in enumerate(cols)])
ax.set_yticklabels(cols)
for i, (rowname, row) in enumerate(moonshot[moonshot['site']==site_type][cols].corr().iterrows()):
for j, (key, val) in enumerate(row.iteritems()):
ax.annotate(f"{val:0.2f}", xy=(i,j), xytext=(-10, -5), textcoords="offset points")
ax.set_title(f"Docking to {site_type}")
Text(0.5, 1.05, 'Docking to active-covalent')
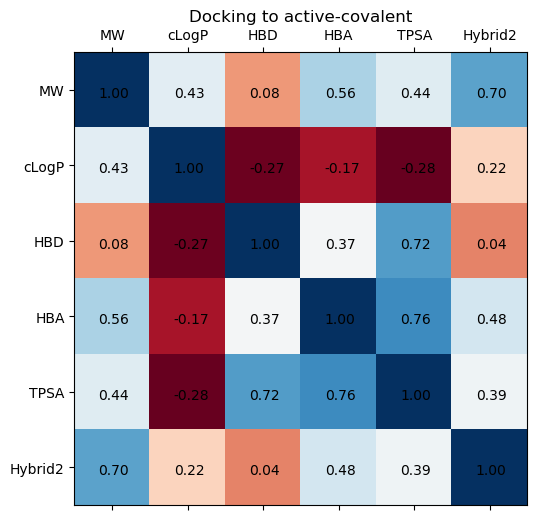
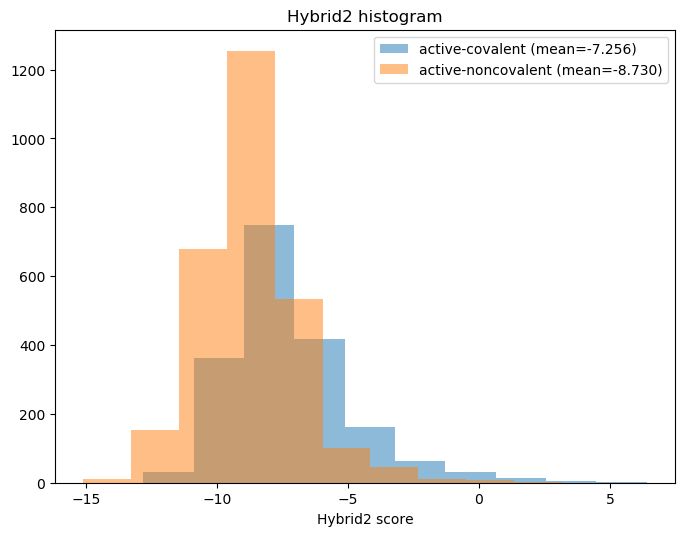
In general, lower docking score seem better, so the noncovalent sites might present more optimal binding locations (see histogram below). This seems non-intuitive because, if active-covalent really means sites that bond covalently, then covalent bonds would seem more energetically favorable than non-covalent interactions. Alternatively, forming covalent bonds might suggest an unstable region of the complex that could be shielded from the surroundings, inhibiting any sort of small molecule from binding the pocket? Expert opinion would be much appreciated here
fig, ax = plt.subplots(1,1, figsize=(8,6), dpi=100)
covalent_mean = moonshot[moonshot['site']=='active-covalent']['Hybrid2'].mean()
noncovalent_mean = moonshot[moonshot['site']=='active-noncovalent']['Hybrid2'].mean()
ax.hist(moonshot[moonshot['site']=='active-covalent']['Hybrid2'], alpha=0.5,
label=f'active-covalent (mean={covalent_mean:.3f})')
ax.hist(moonshot[moonshot['site']=='active-noncovalent']['Hybrid2'], alpha=0.5,
label=f'active-noncovalent (mean={noncovalent_mean:.3f})')
ax.set_title(f"Hybrid2 histogram")
ax.set_xlabel("Hybrid2 score")
ax.legend()
<matplotlib.legend.Legend at 0x7fac6b459850>
from rdkit import Chem
rdkit_smiles = [Chem.MolFromSmiles(a) for a in moonshot.sort_values('Hybrid2', ascending=True)['SMILES'].head(10)]
scores = [f"{a:.3f}" for a in moonshot.sort_values('Hybrid2', ascending=True)['Hybrid2'].head(10)]
img=Chem.Draw.MolsToGridImage(rdkit_smiles,molsPerRow=5,subImgSize=(200,200),
legends=scores)
img

